WSI // ML gets an update
After years of supporting machine learning projects at A.C.Camargo Cancer Center, the annotation software developed during my Ph.D. has received a major update.
After several years of use in supporting various ML projects within the A.C.Camargo Cancer Center institution, I've had the opportunity spend the last few weeks working on an updated version of the WSI // ML software, the annotation software I developed during my Ph.D. This update addresses the evolution of underlying technologies and enhances the software’s role in our ML workflows.
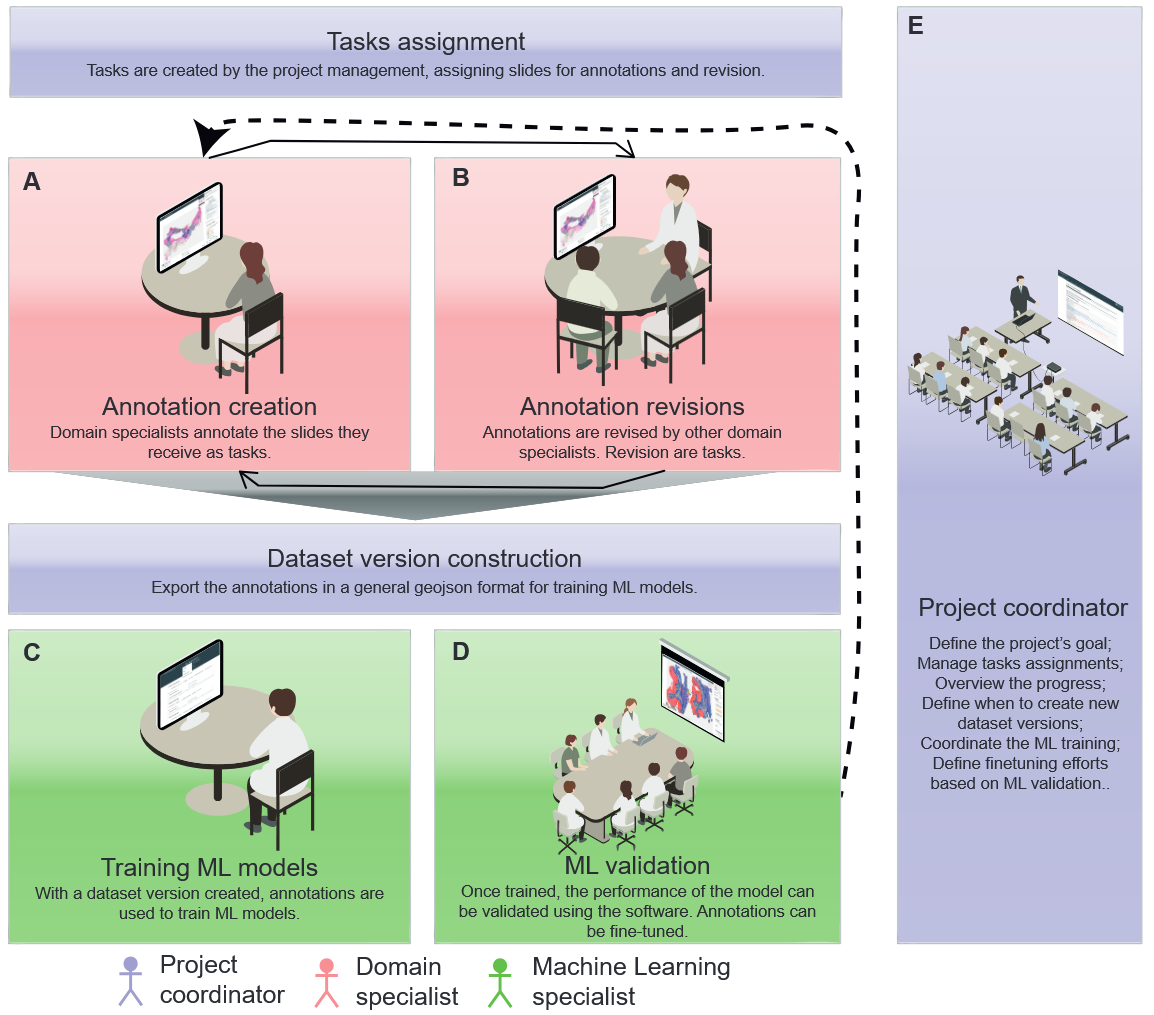
This software implements a framework (Fig. 1) that enables the construction of high-quality histological datasets from large histological images (Whole Slide Images). It streamlines the process by creating clear tasks for pathologists, and it allows project coordinators to easily track annotation progress.
What has changed
This major update brings significant improvements to usability, performance, and functionality. Specifically:
- The web client has been completely rewritten with Vue3, resulting in a more intuitive user interface compared to the first version. We've also improved project organization, making it easier to navigate tasks and manage the datasets. Performance has also been improved, especially when loading multiple slides. The slide listing now displays a thumbnail preview for each image, streamlining the selection process.
- A key feature of this update is the ability to open and annotate multiple slides simultaneously, allowing slides from the same patient to be visualized at the same time. Revision tasks are now better integrated into the workflow and allow for easy visualization of colleague annotations without the need to define a specific revision task, with the same functionality applied to model predictions.
- We've also implemented version control for datasets, allowing you to download previously generated versions whenever needed. Furthermore, you can now upload model annotations directly into the software for comparison and refinement -- previously this was done by the annotator out of the software.
- Under the hood, we've transitioned the backend to MongoDB, replacing the previous SQLAlchemy and PostgreSQL setup for increased flexibility and simplicity. The docker usage has also been revamped: the docker is composed of three containers containing the backend, frontend, and a MongoDB server instance.
Concluding remarks
The ongoing development and use of this software—years after its initial release and the conclusion of several PhD projects at A.C.Camargo—highlights its significant impact on the work of the Cancer Center. I am proud of the journey that took place during my PhD.
