I have concluded my Ph.D.
I am thrilled to share that on 01/04/2025 I have successfully concluded my Ph.D., a process I started back in 2020.
I studied a technique for Graph Neural Network, which we apply to histopathology image segmentation task. We also proposed an annotation software and several annotated datasets.
Histopathology application
The first step was to identify a suitable application to test our proposal. Given our interest in applying the technique to health-related applications, we chose the highly relevant and impactful area of histopathology image analysis, specifically focusing on segmentation tasks. This work was conducted in collaboration with A.C.Camargo Cancer Center.
WSI2ML: annotation software
One of the first challenges we encountered was the lack of machine learning-oriented annotation software that would allow us to delegate tasks efficiently, track annotation efforts systematically, and validate whether our annotation efforts were successfully addressing the task or project objectives we had set out to achieve. To overcome this, we decided to propose our own in-house solution, focusing particularly on enabling the validation of models and improving the dataset through multiple iterations.
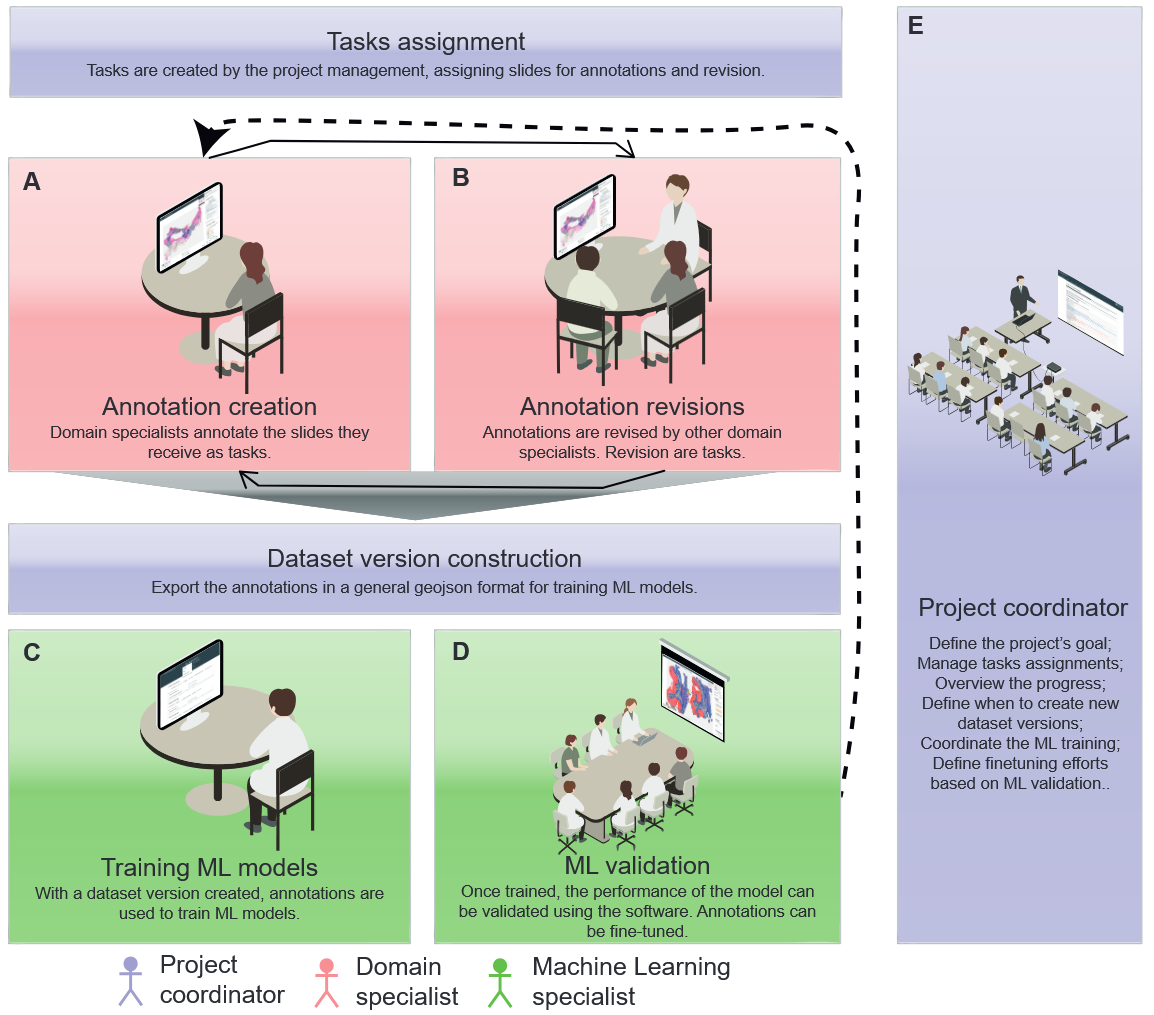
The software was developed using web technologies, including Flask for the backend, SQLAlchemy for database management, and Vue for the user interface. For handling and displaying histopathology slides, we utilized the OpenSlide library, and to handle annotations for large image format, we developed our own annotation library.
The software has been deployed at A.C.Camargo Cancer Center ever since and was crucial in creating several annotated datasets that resulted in publications and other projects within the institution. We published a paper describing the software in WebMedia '23, and the software is also publicly available on GitHub.
Resulting datasets
Over the years, several datasets have been annotated with the proposed tool:
- Gastrointestinal dataset: A dataset created for segmentation tasks in histopathology images of the gastrointestinal tract. I used this dataset to highlight this aspect of our results in my thesis.
- Background and tumor detection dataset for breast tissues: A dataset focusing on background and tumor segmentation in histopathology images of breast tissues. This dataset has played a part in a 2024 publication.
- Cytological dataset: A dataset designed for cytological analysis tasks, aiding in further understanding cellular-level abnormalities in citological images. This dataset is associated with a Ph.D. project that was concluded in 2025.
Activation map-based Graph Neural Network
We also studied a Message Passing-based Graph Neural Network (GNN) method, inspired by discussions with pathologists regarding their experience in analyzing histopathological images. As a result, this method aims to model connectivity patterns by representing the presence or absence of elements of specific features within a defined neighborhood and leveraging a Neural Network to extract meaningful patterns from these representations.
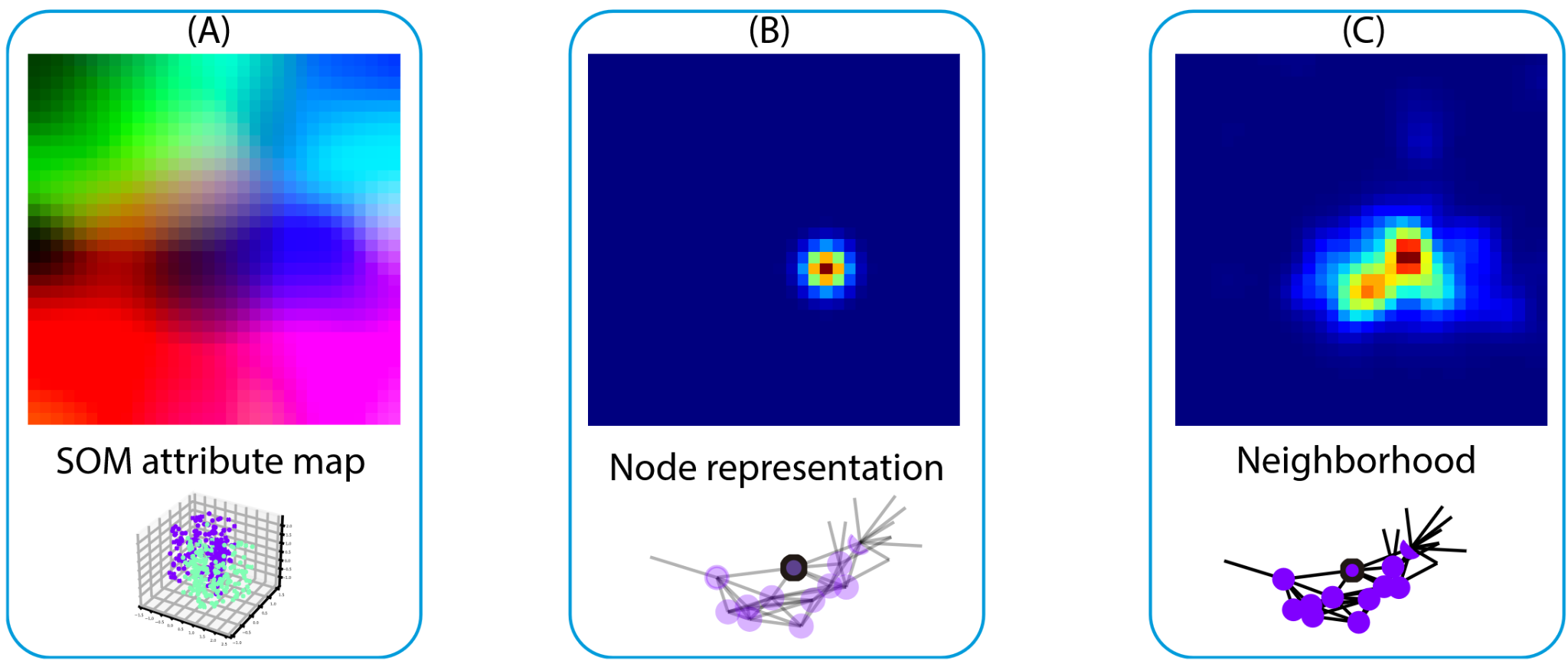
The core idea involves using Self-Organizing Maps (SOM) to project high-dimensional feature data into a lower-dimensional representation. By doing so, we can characterize a neighborhood by identifying and activating regions on this map corresponding to features present in the local neighborhood. This approach enables the network to extract connectivity patterns and relationships based on the spatial and feature context present in histopathology images.
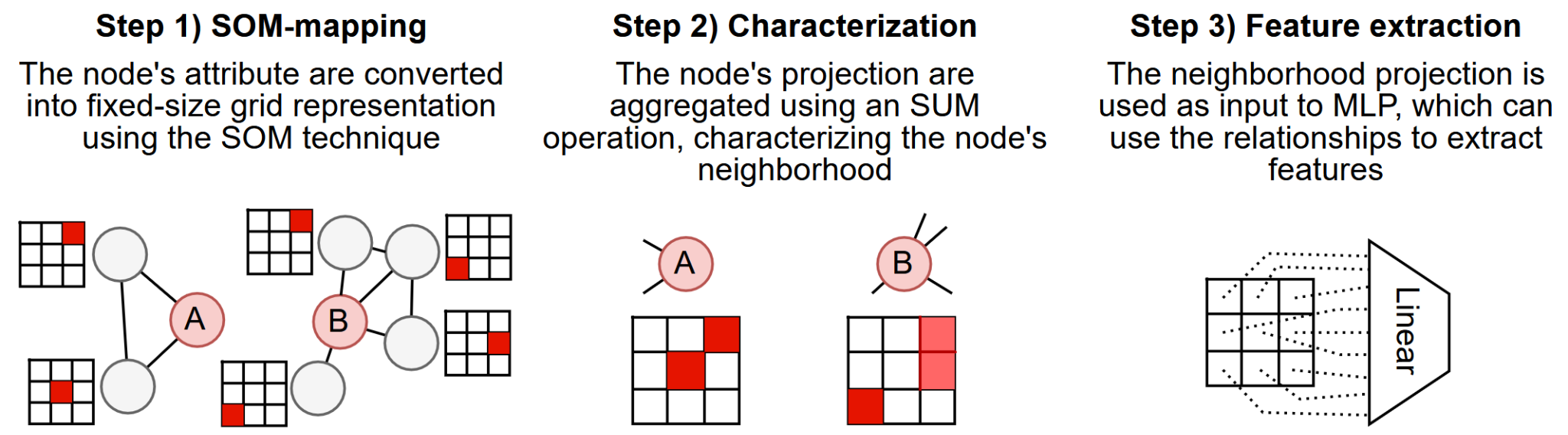
This method enhances interpretability by providing a clear visual representation of the feature activations through the activated map, allowing researchers to understand which features in the local neighborhood contribute to the model's predictions. This combination of spatial and feature context significantly aids in understanding neighborhood relationships while maintaining good performance in tasks that demand such insights, such as histopathological segmentation.
Thesis organization
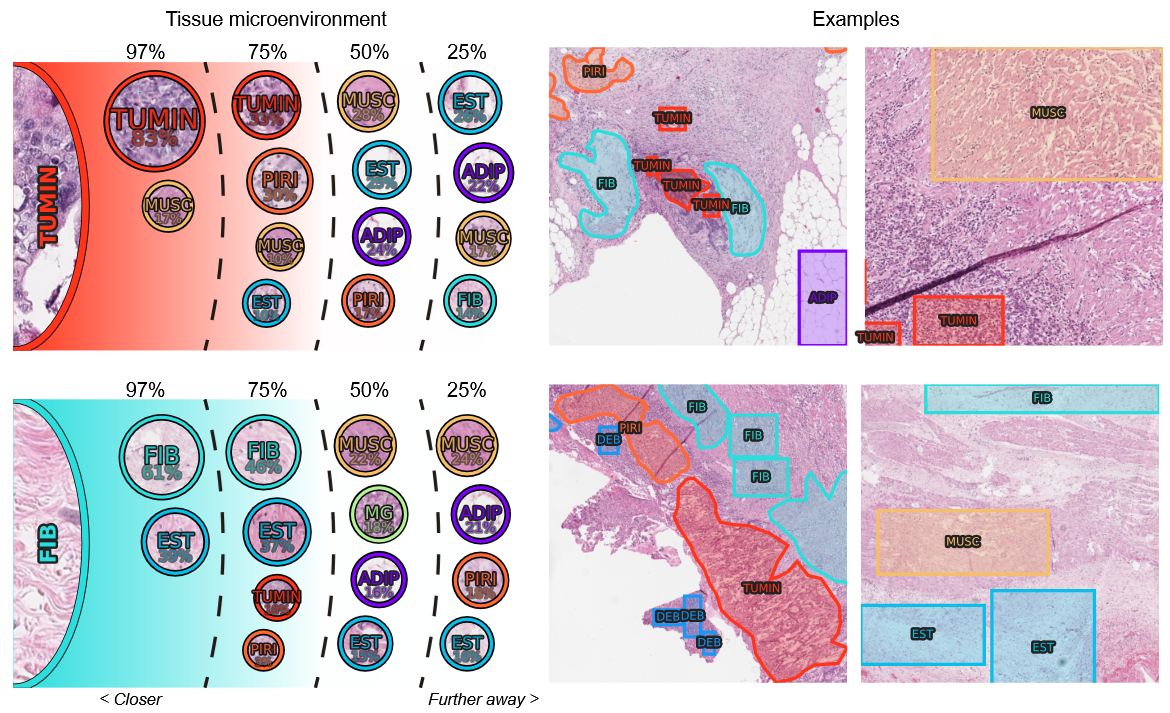
In the thesis, we present the results as they were obtained: in an incremental way. We started with the development of a custom annotation tool designed to meet the specific needs of the histopathological segmentation task. This tool was fundamental in the creation of high-quality annotated datasets, from which several were created, and we highlight the Gastrointestinal dataset. We complete the cycle by training the GNN method on that dataset, demonstrating the interdependence between the tool, the data, and the model's performance. Finally, we use the advantages to interpretability that the method grants to understand the patterns and contrast them to a pathologist, which validated the patterns the model learned.
Resulting publications
Throughout my Ph.D. journey, I had the privilege of contributing to multiple publications that stemmed from the research and developments carried out during this period. Below, I've listed some of the publications that resulted from this work:
An Interpretable Activation Map-based Graph Neural Network for Histological Image Classification
Martins, L., Bueno, A.P., Valieris, R., Defelicibus, A., Da Silva, I.T. and Zhao, L. Pre-print.
